Scraping Corona data from the Norwegian Directorate of Health and the Norwegian Institute of Public health websites
Disclaimer. This post has simply been a personal exercise in webscraping data that others have made available. I am also personally curious to see the correspondence between the observed numbers of patients being hospitalized and in ICU and the predictions given the different RE-estimates produced.
This project uses daily updates about patients hospitalized and in ICU scraped from the Norwegian Directorate of Health in tandem with the predictions from the report “Covid-19 epidemic: Risk assessment and response in Norway”1 published by the Norwegian Institute of Public Health (NIPH) building on modelling conducted by the Imperial College COVID-19 Response Team.
I have resorted to web-scraping as I have not been able to find suitable data published online. Please advice if you now of any open datasources containing this information!
The figure plots the daily number of observed cases (the red dot) on the timeline of predictions from the report from the NPHI (the red, green and blue lines represent trajectories predicted from different REs). This far, the observed numbers are lagging behind the predictions and will only be in sync from March 30th and onwards.
Note. The report from the NIPH provided numeric estimates (with confidence intervals) for the dates March 23rd (observed data), March 30th, April 6th and April 13th. The data in between these dates is based on a linear prediction based on the trends from the estimates between these dates. This was done in Excel as the data was manually entered from the NIPH-report which was published as a pdf.
Update 1. I have had to update the scraping script due to the Norwegian Directorate of Health adding an extra space (" ") in front of the number of patients on ventilators on March 29th thus breaking the method of using strsplit and retrieving the object from a specific position (which shifted the number is wanted from position [14] to position [15].
Update 2. The number of intensive care patients reported by the Norwegian Directorate of Health are the numbers in need of ventilators. I have therefore updated the script - possibly even more fragile - reading the numbers of the website of the Norwegian Institute of Public health. This requires some hacky attempt to foresee the date included in the website, and there are problems as this date sometimes adds the weekday and the date, and other times only the date. It also parses numbers from the text printed on the website, so adding a word or reordering the numbers will make the script fail miserably… the only available source of these numbers seem to be either this website, some newspapers or the official registers that I don’t have access to.
library(readxl)
library(dplyr)
library(ggplot2)
library(hrbrthemes)
source("corona.R") # See scraping script below
hdir_obs <- as.data.frame(
read.table("innl_date.csv",
col.names = c("row", "day", "month", "year", "innlagt", "intensiv", "date")
)
)
hdir_obs$date <- as.POSIXct(hdir_obs$date, format="%d.%m.%Y", tz="UTC")
fhi_obs <- as.data.frame(
read.table("inf_date.csv",
col.names = c("row", "day", "month", "year", "infected", "date", "day_tmp", "mth_tmp")
)
)
fhi_obs$date <- as.POSIXct(fhi_obs$date, format="%d.%m.%Y", tz="UTC")
fhi_innl <- as.data.frame(
read.table("infint_fhi.csv",
col.names = c("row", "day", "month", "year", "infected_fhi", "intensiv_fhi", "date")
)
)
fhi_innl$date <- as.POSIXct(fhi_innl$date, format="%d.%m.%Y", tz="UTC")
fhi_innl <- fhi_innl %>% select(-row, -day, -month, -year)
coronoa_pred <- read_excel("coronoa_pred.xlsx")
corona_pred_obs <- full_join(coronoa_pred, hdir_obs, by = "date")
corona_pred_obs <- corona_pred_obs %>% select(-row, -day, -month, -year)
corona_pred_obs <- left_join(corona_pred_obs, fhi_innl, by = "date")
corona_pred_obs <- corona_pred_obs %>% mutate(re = as.factor(re))corona_pred_obs %>%
ggplot(aes(x=date, y=intensiv_pred, ymax= intensiv_upr, ymin = intensiv_lw, group = re, fill = re)) +
geom_ribbon() +
geom_line(lty="dashed") +
geom_point(data = corona_pred_obs, aes(x=date, y = intensiv_fhi), color = "red") +
theme_ipsum_rc() +
labs(main = "Intensive care patients", x = "Date", y = "Predicted number of \nintensive care patients",
caption="Data from the Norwegian Institute of Public Health")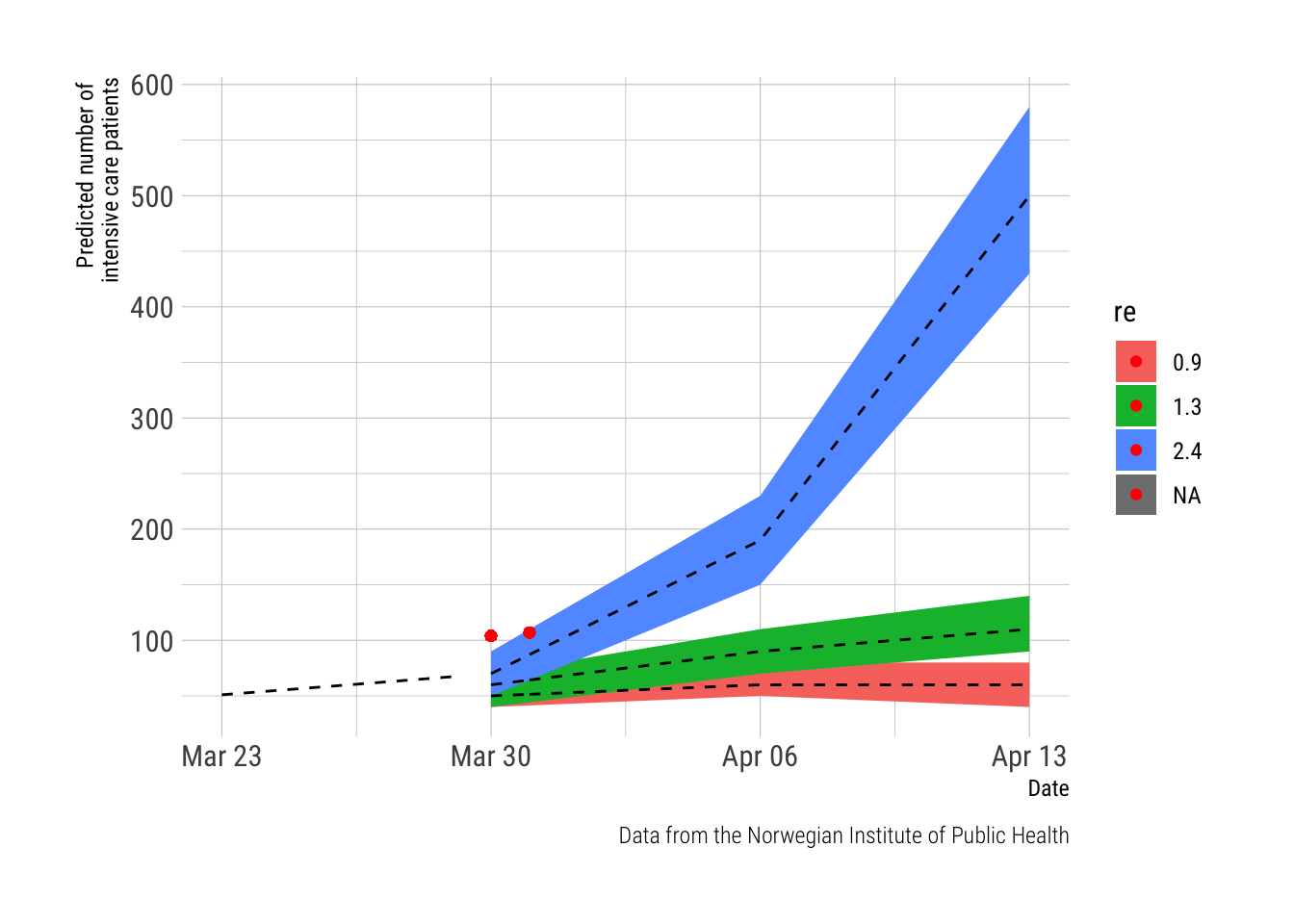
corona_pred_obs %>%
ggplot(aes(x=date, y=innlagte_pred, group = re, ymin = innlagte_lw, ymax = innlagte_upr, fill = re)) +
geom_ribbon() +
geom_line(lty="dashed") +
geom_point(data = corona_pred_obs, aes(x=date, y = innlagt), color = "red") +
theme_ipsum_rc() +
labs(main = "Hospitalized patients", x = "Date", y = "Predicted number of \nhospitalized patients",
caption="Data from the Norwegian Directorate of Health and the Norwegian Institute of Public Health")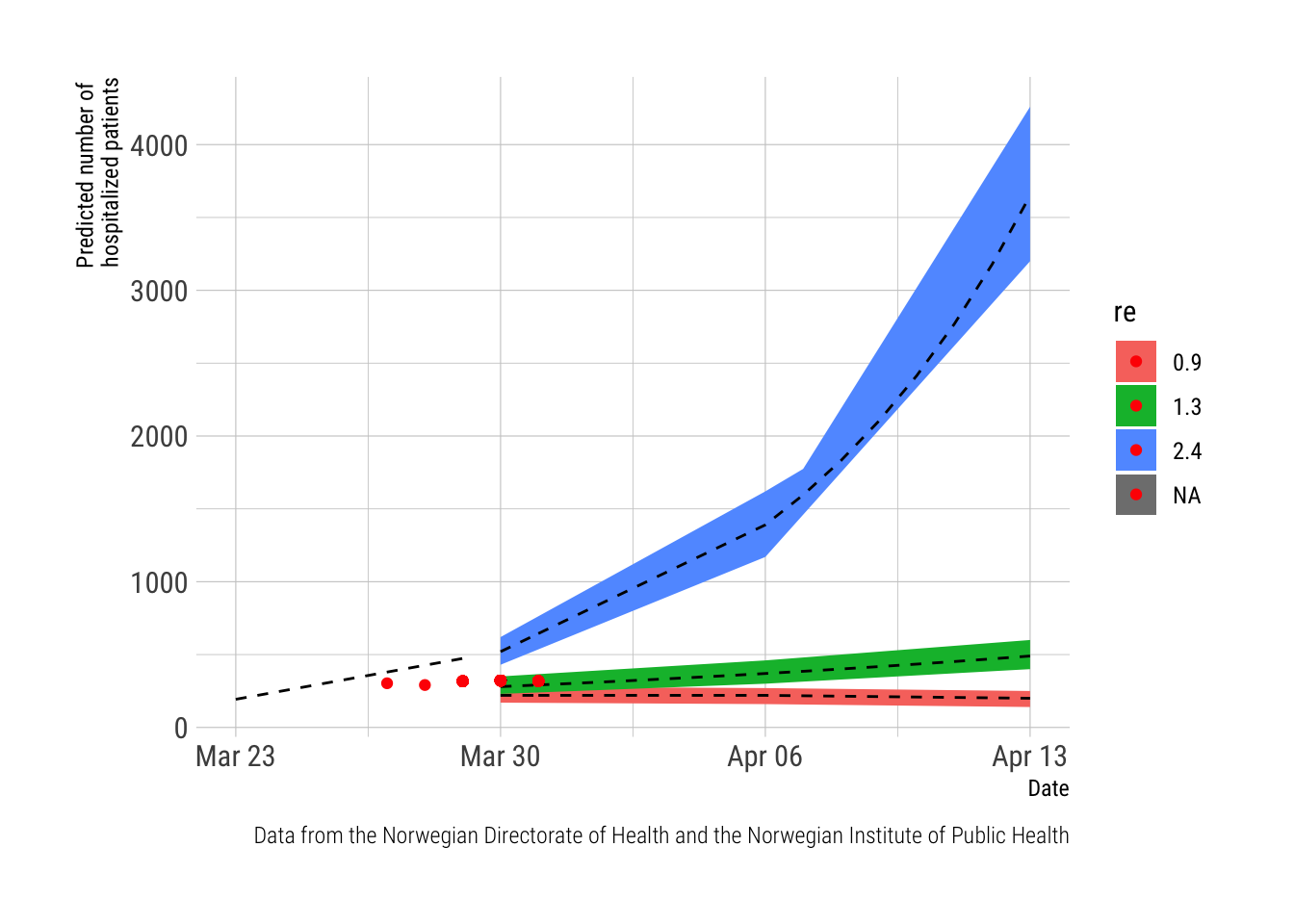 The script to scrape the websites is documented below. It does the scraping and stores results to two csv-files. The script is sourced by this post, to keep it updated and the resulting csv files are read into this script and used to produce the graphs. For now, this requires a manual update of the script to keep it current, and I have not investigated options to make the script self-updated, which could probably be achieved with shiny or other options. The script to do this is very fragile; It depends on the Norwegian Directorate of Health keeping the address to their site and even the wording on their site exactly similar from day to day. I have already had to update it due to the Norwegian Directorate of Health adding an extra space (" ") in front of the number of ICU patients on March 29th thus breaking the method of using
The script to scrape the websites is documented below. It does the scraping and stores results to two csv-files. The script is sourced by this post, to keep it updated and the resulting csv files are read into this script and used to produce the graphs. For now, this requires a manual update of the script to keep it current, and I have not investigated options to make the script self-updated, which could probably be achieved with shiny or other options. The script to do this is very fragile; It depends on the Norwegian Directorate of Health keeping the address to their site and even the wording on their site exactly similar from day to day. I have already had to update it due to the Norwegian Directorate of Health adding an extra space (" ") in front of the number of ICU patients on March 29th thus breaking the method of using strsplit and retrieving the object from a specific position (which shifted the number is wanted from position [14] to position [15]. The data is usually publised on the Norwegian Directorate of Health website around 13:00 (1 pm.).
library(tidyverse)
library(rvest)
library(xml2)
today <- format(Sys.time(), "%a %b %d %X %Y")
day <- unlist(strsplit(today, " "))[1]
day <- recode(day,
Mon = "mandag",
Tue = "tirsday",
Wed = "onsdag",
Thur = "torsdag",
Fri = "fredag",
Sat = "lordag",
Sun = "sondag")
mth <- unlist(strsplit(today, " "))[2]
mth <- recode(mth,
Mar = "mars",
Apr = "april")
date <- unlist(strsplit(today, " "))[3]
year <- unlist(strsplit(today, " "))[5]
url3 <- paste0("https://www.fhi.no/nyheter/2020/status-koronasmitte-", day, "-", date, ".-", mth, "-", year)
url_res3 <- xml2::read_html(url3)
frafhi <- url_res3 %>% html_nodes(".textual-block") %>%
map_df(~{
tibble(
# postal = html_node(.x, "span") %>% html_text(trim=TRUE),
fhi = html_nodes(.x, "ul > li") %>% html_text(trim=TRUE)
)
})
innlagt_fhi <- frafhi$fhi[1] %>% str_match_all("[0-9]+") %>% unlist %>% as.numeric
intensiv_fhi <- frafhi$fhi[4] %>% str_match_all("[0-9]+") %>% unlist %>% as.numeric
innl_num <- word(innlagt_fhi)[1:2]
innl_num <- as.numeric(paste0(innl_num[1], innl_num[2]))
intens_num <- as.numeric(word(intensiv_fhi)[3])
# month <-
fulldate <- format(Sys.time(), "%d.%m.%Y")
fullmonth <- format(Sys.time(), "%m")
fullyear <- format(Sys.time(), "%Y")
infint_fhi <- as.data.frame(cbind(date, fullmonth, fullyear, innl_num, intens_num, fulldate))
write.table(infint_fhi, "/Users/st06810/Dropbox/UiB/blog/content/post/infint_fhi.csv", dec=".", quote=FALSE, append=T, sep="\t", col.names=FALSE)
url2 <- paste0("https://www.helsedirektoratet.no/nyheter/oversikt-over-innlagte-pasienter-med-covid-19-per-", format(Sys.Date(), "%d"),".mars")
url_res2 <- xml2::read_html(url2)
url_res2 %>% html_text()
hdir <- url_res2 %>%
html_nodes(".b-article-intro__intro") %>% html_text()
day_hdir <- unlist(strsplit(hdir, " "))[3]
day_hdir <- gsub('[[:punct:] ]+','', day_hdir)
mth_hdir <- unlist(strsplit(hdir, " "))[4]
mth_hdir <- ifelse(mth_hdir=="mars", "03",
ifelse(mth_hdir=="april", "04",
ifelse(mth_hdir=="mai", "05",
ifels(mth_hdir=="juni", "06", NA))))
n_innl_hdir <- unlist(strsplit(hdir, " "))[6]
n_int_hdir <- unlist(strsplit(hdir, " "))[15]
# This function is supposed to replace NULL values with missing values in case the script is used
# before the website is updated. Not working properly yet...
if (is.null(day_hdir) && is.null(day_hdir && is.null(mth_hdir) && is.null(n_innl_hdir) && is.null(n_int_hdir))){
day_hdir <- NA
mth_hdir <- NA
n_innl_hdir <- NA
n_int_hdir <- NA
return(day_hdir)
return(mth_hdir)
return(n_innl_hdir)
return(n_int_hdir)
}
# day_hdir
# mth_hdir
# n_innl_hdir
# n_int_hdir
year_hdir <- 2020
date <- (paste(day_hdir, mth_hdir, year_hdir, sep = "."))
innl_date <- as.data.frame(cbind(day_hdir, mth_hdir, year_hdir, n_innl_hdir, n_int_hdir, date))
write.table(innl_date, "/Users/st06810/Dropbox/UiB/blog/content/post/innl_date.csv", dec=".", quote=FALSE, append=T, sep="\t", col.names=FALSE)
url <- "https://www.fhi.no/sv/smittsomme-sykdommer/corona/"
url_res <- xml2::read_html(url)
n_inf <- url_res %>%
html_nodes(".fhi-key-figure-number") %>% .[[2]] %>% html_text()
n_inf <- as.numeric(n_inf)
date_txt <- url_res %>%
html_nodes(".fhi-key-figure-desc") %>% .[[2]] %>% html_text()
day_tmp <- unlist(strsplit(date_txt, " "))[7]
mth_tmp <- unlist(strsplit(date_txt, " "))[8]
day <- gsub('[[:punct:] ]+','',day_tmp)
day <- as.numeric(day)
mth <- ifelse(mth_tmp=="mars", "03",
ifelse(mth_tmp=="april", "04",
ifelse(mth_tmp=="mai", "05",
ifels(mth_tmp=="juni", "06", NA))))
year <- 2020
date <- (paste(day, mth, year, sep = "."))
mth <- as.numeric(mth)
inf_date <- as.data.frame(cbind(day, mth, year, n_inf, date, day_tmp, mth_tmp))
write.table(inf_date, "/Users/st06810/Dropbox/UiB/blog/content/post/inf_date.csv", dec=".", quote=FALSE, append=T, sep="\t", col.names=FALSE)Covid-19-epidemien: Risiko, progonose og respons i Norge etter uke 12. Med vedlegg. 24.3.2020↩︎